The way everyone has been tracking and talking about viral variants of COVID-19 has been quite interesting. The start of the pandemic was a live-lab teaching moment for everyone on epidemiology. Then as the first vaccines were released, the world learned about vaccines, RNA, and viral molecular biology. Now, as active variants have been named, folks are learning about evolution, mutations, and the viral lifecycle.
As a scientist at heart, I’ve found all of it quite invigorating.
Making with really small tools
I’ve always had an interest in the structure of DNA and proteins. Long before I picked up a soldering iron, I was splicing DNA and building bespoke molecules.
As a technician at MIT, what little research I did was focused on characterizing some features of repetitive DNA. I was fortunate enough to be given the opportunity to learn some new techniques for injecting defined sequences of DNA into fruit flies, to modify their genome.
In grad school, I had wanted to do my rotation thru a microbiology lab to do some fun evolutionary studies using microbes. Alas, that professor was away, so instead I used electron microscopes to peer into and measure DNA at its smallest scales.
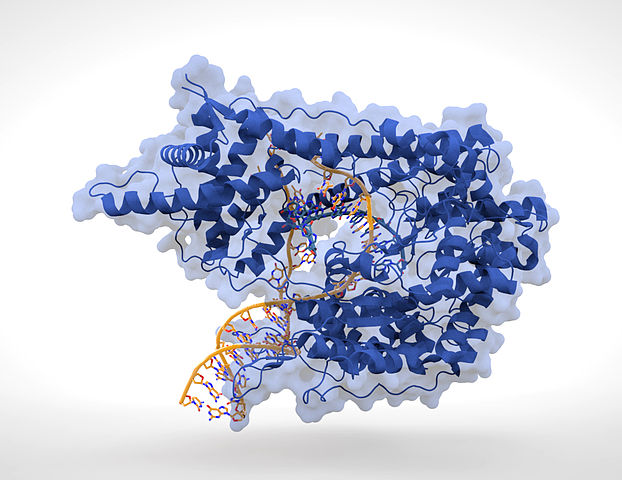
For my thesis work, I took my interest in DNA and proteins a step further. Our lab studied DNA-protein binding. We’d make DNA with modifications that effectively changed it by one or a few atoms. We then had a very simple model system (using RNA and T7 polymerase, if you care to know, shown in the image above) to read out the effects of these atomic-level changes – how it affected the binding characteristics, how it affected the activity of the enzyme we used.
True molecular biology.
Benchtop synthesizer
One of the cool aspects of this work was that we had our own DNA synthesizer, a tool normally meant for big core facilities making oodles of DNA.
Our model was a small one (sat on one end of my bench) with which we could make our short polymers of DNA to spec. In particular, we would alter the process to add modified bases, bought or synthesized in our lab, so that we were not stuck at the traditional A, C, G, and T. We also did some other crazy things to the DNA to modify it in various ways – all at the molecular level.
Making proteins
For my post-doc, I headed to a genetics lab, where were were mapping chromosomes and discovering new genes. Though, it so happened that one of the genes was an enzyme inhibitor, and, being the resident biochemist, I was asked to characterize the inhibitor.
Just like I did modifications to the DNA, I made modifications to the protein. While the chemistry of synthesizing proteins is way complex (no, I didn’t have a synthesizer), the tools of molecular biology (using bacteria) were sufficient for me to vary the amino acids across the inhibitor to alter its function. Due to the variety of natural amino acids, I could alter my inhibitor to create variants that differed by an atom or a few atoms.
And using some fun biochemistry (and fluorescent peptides – see, blikenlights even then) I was able to characterize what the inhibitor could do (FWIW, it was inhibiting enzymes from with different class of chemistry, blah blah).
Making at the molecular level
This was all long ago, and was a class of magic I took for granted. All the recent talk of viral epidemiology, structure, and immunology brought back memories of those years and reminded me that way back then, I was a maker too. A maker using really small tools and really small builds. Haha.
Not surprised I’m still a maker.
Image: Thomas Splettstoesser